Afr. J. Parasitol. Mycol. Entomol. 2024, 2(1), 1; doi:10.35995/ajpme2010001
Performance of Chromogenic Candida Lab-Agar® Medium in Presumptive Identification of Candida Species from Clinical Samples at Sourô Sanou University Hospital of Bobo-Dioulasso, Burkina Faso
1
Ecole Doctorale des Sciences de la Santé, Université Nazi-BONI, 01 BP 1091 Bobo-Dioulasso, Burkina Faso; yerbis2000@yahoo.fr (I.W.Y.); badobassabdulel@gmail.com (B.B.); isidore.mandy@gmail.com (I.M.); hsanata@yahoo.fr (S.B.)
2
Centre Muraz/ Institut National de Santé Publique, 01 BP 390 Bobo-Dioulasso 01, Burkina Faso
3
Pôle de Microbiologie Médicale, Institut de Recherche Expérimentale et Clinique (IREC), Université catholique de Louvain (UCLouvain), Bruxelles, Belgique; ahalieyah.anantharajah@student.uclouvain.be (A.A.); hector.rodriguez@uclouvain.be (H.R.-V.)
4
Service des laboratoires, Centre Hospitalier Universitaire Régional de Ouahigouya, 01 BP 36 Ouahigouya 01, Burkina Faso
5
Departement de Microbiologie, Cliniques Universitaires Saint-Luc, Université catholique de Louvain, Avenue Hippocrate 10, 1200, Bruxelles, Belgique
6
Department of Microbiology, CHU Namur site-Godinne, Université Catholique de Louvain, Rue Dr Gaston Therasse 1, 5530 Yvoir, Belgium; carlotamonher@gmail.com (I.M.); odenis@ulb.ac.be (O.D.)
7
Ecole de Santé Publique, Université Libre de Bruxelles, Brussels, Belgium
8
Département des laboratoires, Centre Hospitalier Universitaire Sourô Sanou, 01 BP 676 Bobo Dioulasso 01, Burkina Faso
*
Corresponding author: naksaid2006@yahoo.fr; Tel.: +226-76139791
How to cite: Nakanabo Diallo, S.; Yerbanga, I.W.; Bado, B.; Mandy, I.; Anantharajah, A.; Montesinos, I.; Denis, O.; Bamba, S.; Rodriguez-Villalobos, H. Performance of Chromogenic Candida Lab-Agar® Medium in Presumptive Identification of Candida Species from Clinical Samples at Sourô Sanou University Hospital of Bobo-Dioulasso, Burkina Faso. Afr. J. Parasitol. Mycol. Entomol., 2024, 2(1): 1; doi:10.35995/ajpme2010001.
Received: 11 August 2023 / Accepted: 6 December 2023 / Published: 3 January 2024
Abstract
:Introduction: The incidence of Candida infections is increasing worldwide. In clinical laboratories of resource-constrained countries, Candida speciation is commonly limited to germ tube tests and culture onto a chromogenic medium. In this study, we evaluated the diagnostic performance of Chromogenic Candida Lab-Agar® (CCL) in identifying Candida species from clinical samples. Methods: We evaluated the diagnostic performance of CCL with 83 yeast isolates collected from 73 clinical samples at the laboratory department of Sourô Sanou University Hospital of Bobo-Dioulasso, Burkina Faso. Clinical specimens included vaginal swabs, urine, and blood cultures. After preliminary isolation on Sabouraud chloramphenicol agar, yeast isolates were inoculated onto the CCL medium and incubated at 35 °C for 48 h. Matrix-assisted laser desorption/ionisation time of flight mass spectrometry (MALDI-TOF MS) and ribosomal DNA internal transcribed spacer (ITS) sequencing were used as reference methods. Results: Among yeast species, Candida albicans was the most prevalent (43.4%), followed by C. krusei (13.3%), C. glabrata (12.0%), C. kefyr (8.4%), and C. tropicalis (7.2%). The overall agreement rate of CCL was 56.6% and varied across Candida species; it was 94.4% for C. albicans, 50% for C. glabrata, 18.2% for C. krusei, and 33.3% for C. tropicalis. Conclusions: This study showed that CCL had moderate accuracy in identifying Candida at the species level from clinical specimens in a routine laboratory in Burkina Faso. The misidentification of non-albicans species may expose patients to inadequate antifungal treatment. Therefore, identifying yeast in a routine based on CCL is not enough and should be associated with more accurate methods.
Keywords:
Candida species; chromogenic medium; performance; Bobo-Dioulasso1. Introduction
Candidiasis is an infection due to fungi of the Candida genus and constitutes a significant cause of morbidity and mortality in humans [1]. Candida albicans is the most common yeast identified in clinical samples. However, recent studies showed a rising proportion of non-albicans Candida species worldwide, including C. glabrata, C. krusei, C. tropicalis, and C. parapsilosis [1,2,3,4,5]. Therefore, candidiasis management is more and more challenging due to the geographical diversity of Candida species and their variable profile of susceptibility against antifungal agents [6,7,8,9,10]. The identification of Candida at the species level in clinical samples is, therefore, a prerequisite for the therapeutic decisions and guidance of local strategies in the prophylaxis and empirical treatment of candidiasis [11].
In most clinical laboratories in resource-limited settings, the diagnosis of Candida species relies upon germ tube testing, culture on chromogenic medium, or conventional biochemical testing. The chromogenic agar medium is one of the common methods used especially since it is cost-effective, easy to use, and allows the presumptive identification of the main pathogenic Candida species [12,13,14].
In the current study, we analysed the performance of Chromogenic Candida Lab-Agar® (BioMaxima Biocorp, Warsaw, Poland), used in routine practices in the laboratory of the Sourô Sanou University Hospital, for the identification of pathogenic yeast at the species level. MALDITOF-MS and DNA ITS sequencing were considered reference methods.
2. Methods
2.1. Strain Collection
We analysed a collection of 83 yeast strains isolated from 73 clinical samples at the laboratory department of Sourô Sanou University Hospital in Bobo-Dioulasso, Burkina Faso. Clinical specimens were obtained during a five-month period (January to May 2021) and were from various clinical origins, including vaginal swabs (45), urine (27), and blood (1). Clinical samples were first streaked on Sabouraud Dextrose Agar containing chloramphenicol (SDA) and incubated at 35 °C for 48 h. Yeast-like colonies yielded on SDA were subsequently identified on the CCL medium.
2.2. Chromogenic Medium Identification
The chromogenic Candida lab-agar® medium (BioMaxima Biocorp, Warsaw, Poland) was prepared from dehydrated powder, in accordance with the manufacturer’s instructions. Agar plates were stored at 2–8 °C and equilibrated at room temperature before use.
Yeast-like colonies on SDA were inoculated onto CCL plates by streaking them with sterile plastic loops. Inoculated plates were then incubated at 35 °C for 48 h. Candida species identification was performed based on the visual observation of the colour and morphology of the yeast colonies in accordance with the manufacturer’s guidelines. Isolates showing atypical colours were considered undetermined species. Chromogenic Candida Lab-Agar® allowed the identification of four Candida significant species (C. albicans, C. glabrata, C. krusei, and C. tropicalis), as shown in Figure 1. After CCL identification, yeast isolates were aliquoted in distilled water and stored at −20 °C.
2.3. Reference Methods
The yeast isolates were secondarily identified at the microbiology department of Cliniques Universitaires Saint-Luc, UCLouvain (Brussels, Belgium) using matrix-assisted laser desorption ionisation time of Flight (MALDI-TOF) mass spectrometry (Brucker Daltoniks, Bremen, Germany) and DNA sequencing.
MALDI-TOF-mass spectrometry identification: Fresh colonies were obtained from aliquots via culturing on Sabouraud chloramphenicol and a gentamicin dextrose agar plate (Dickinson, Sparks, MD, USA) and incubated for 24 h at 35 °C. A single colony was directly spotted onto the MALDI-TOF target plate and overlaid with 1 μL of 70% formic acid. As soon as the spot dried, 1 μL of a matrix (α-cyano-4-hydroxycinnamic acid) was applied and allowed to dry at room temperature. Then, the target plate was placed in the MALDITOF ionisation chamber. The spectra were analysed using the Microflex™ LT/SH smart system with MBT Compass IVD software plus the MBT IVD library. An identification score of 1.7 or above was accepted as a reliable result at the species level. Isolates showing lower scores were re-identified after the performance of the ethanol–formic acid extraction method in accordance with the manufacturer’s recommendations. Isolates that failed to be identified via MALDI-TOF (score <1.7) were finally identified via DNA sequencing.
DNA sequencing: DNA extraction was carried out from lysed yeast aliquots using an automated DNA extractor Qiasymphony® SP (QIAGEN, Carlsbad, CA, USA). The internal transcribed spacer regions (ITS1-5.8s-ITS2) of ribosomal DNA were amplified via PCR using specific primers, ITS4 (5′ TCC-TCC-GCT-TAT-TGA-TAT-GC-3′) and ITS5 (5′-GGA-AGT-AAA-AGT-CGT-AAC-AAG-G-3′) [15]. The thermocycling program used was initial denaturation (at 95 °C for 10 min), 30 cycles of denaturation (at 95 °C for 30 s), annealing (at 60 °C for 30 s) and elongation (at 72 °C for 1 min), and then a stabilisation (at 72 °C for 7 min).
The PCR product was preliminarily run on 3% agarose gel and purified using the Illustra ExoProstar® kit (GE Healthcare UK). Sanger sequencing of the amplicons was performed using the Big Dye® Terminator v3.1 Cycling sequencing kit with the following conditions: initial denaturation (at 96 °C for 1 min), 25 cycles of denaturation (at 96 °C for 10 s), annealing (at 50 °C for 5 s), elongation (at 60 °C for 4 min), and then stabilisation at 10 °C for up to 24h. The sequences were purified through Sephadex® G50 DNA Grade and sequence analysis was performed on 3500xL Dx Genetic Analyzer® (Applied Biosystems, Foster City, CA, USA). The obtained sequence data were interpreted using bioinformatic software (Geneious Prime®, Version 2021.2.2) and fungal databases “http://www.mycobank.org, accessed on 07 September 2021)”.
2.4. Data and Statistical Analysis
Identified strains on the CCL medium were categorised as C. albicans, C. krusei, C. glabrata, C. tropicalis, and undetermined (for atypical colours due to the presence of yeast species other than the four mentioned).
Chromogenic Candida Lab-Agar® diagnostic performance was determined through the calculation of the overall agreement between CCL and reference methods, and for each species, we determined the sensitivity, specificity, and positive and negative predictive value.
Calculations and statistical analysis were performed using Microsoft Excel (Microsoft corporation, Redmond, WA, USA).
2.5. Administrative Authorisation
Administrative authorisation was obtained from Sourô Sanou University Hospital authorities prior to yeast strain collection.
3. Results
3.1. Distribution of Yeast Species
The distribution of all 83 yeast isolates according to the clinical source is shown in Table 1. Reference methods allowed identification of 13 species belonging to three genera: Candida, Cyberlindnera, and Trichosporon. Among the yeast species, C. albicans was the most prevalent (43.37%). Overall, non-albicans Candida strains accounted for 51.81% of isolates, with a predominance of C. krusei (13.25%), C. glabrata (12.05%), C. kefyr (8.43%), and C. tropicalis (7.22%). The four Candida species identifiable on CCL represented 75.90% of the study sample. Two Trichosporon spp. were recovered from urine and genital swab. MALDITOF-MS allowed the speciation of all the strains except five, which were identified using DNA sequencing. The strains identified via DNA sequencing included Meyerozyma caribbica (anamorph Candida fermentati) and Diutina mesorugosa (anamorph Candida mesorugosa).
3.2. Candida Species Identification on CCL Medium
The chromogenic Candida Lab-Agar® medium allowed the identification of 72/83 (86.7%) yeast isolates at the species level (Table 2). Almost all C. albicans isolates were correctly identified using the CCL medium except for two isolates. A green to blue-green colony colour on the CCL medium (specific to C. albicans) was also encountered with other species including C. tropicalis (n = 4), C. glabrata (n = 3), Trichosporon spp. (n = 2), C. kefyr (n = 2), C. fermentati (n = 1), or C. krusei (n = 1). Twelve isolates including C. krusei (n = 4), C. kefyr (n = 2), Cyberlindnera fabianii (n = 2), C. fermentati (n = 2), C. guilliermondii (n = 1), and C. lusitaniae (n = 1) were misidentified as C. glabrata on the CCL medium. Three isolates of C. kefyr and one of C. glabrata were incorrectly identified as C. krusei using the CCL medium.
3.3. Chromogenic Candida Lab-Agar® Performance
The overall agreement rate between the CCL medium and reference methods was 56.63% (47/83). The performance of CCL varied among Candida species. Compared to the reference methods, the sensitivity and specificity of the CCL medium for the detection of C. albicans were 94.44% and 72.34%, respectively. Chromogenic Candida Lab-Agar® sensitivity values in the identification of non-albicans Candida species were 50%, 18.18%, and 33.33% for C. glabrata, C. krusei, and C. tropicalis, respectively (Table 3).
4. Discussion
This study evaluated the diagnostic performance of Chromogenic Candida Lab-Agar® media (BioMaxima Biocorp, Warsaw, Poland) in identifying the most prevalent Candida spp. from clinical samples in routine laboratory work in Burkina Faso.
Several methods are used to identify Candida at the species level in clinical laboratories. Progressively, classic morphological and biochemical methods are being replaced by new ones based on molecular or proteomic approaches, allowing more rapid and more accurate identification [12,16]. For routine use in clinical microbiology laboratories, the choice of method for identifying yeast species depends on several factors including the affordability and reliability of the test, and the time consumed [17]. In most clinical laboratories of resource-rich countries, MALDI-TOF MS became the routine method and gold standard for bacterial and fungal identification due to its ease of use, rapidity, and accuracy [18,19]. However, the investment required for its equipment, software, and maintenance constrains its access to resource-limited settings, where other diagnostic tools such as the germ tube test, and subculture on chromogenic media are still the most commonly used methods in routine work. Indeed, chromogenic media are a simple, lower-cost method allowing the diagnosis of the major pathogenic Candida species [20]. On chromogenic media, Candida spp. are identified via the colour and appearance produced through the interaction of the chromogenic substrate and species-specific enzymes secreted by the yeast when growing on the medium. There are many commercially available chromogenic media for Candida identification, with variable performance depending on the manufacturer, strain origins, and experimental conditions. Overall, these media showed variable diagnostic performance with sensibilities and specificities ranging from 49.6 to 100% and 56.3 to 100%, respectively [21,22,23,24,25].
To the best of our knowledge, this study was the first of its kind of the analysis performed evaluating the diagnostic performance of the CCL medium in a routine clinical laboratory setting.
Our results showed a moderate performance of CCL (56.63%) for the identification of Candida species at the clinical laboratory of the Sourô Sanou University Hospital (Bobo-Dioulasso). Indeed, the sensitivity and specificity of the CCL medium in detecting C. albicans are good and concordant with the performances of other chromogenic media. In contrast, concerning non-albicans Candida species including C. glabrata, C. tropicalis, and C. krusei, the sensitivity of the CCL medium is inferior that in the data from the literature [22,26,27].
In routine practices, the difference in colony colours between Candida species was less striking than mentioned by the manufacturer. The expected species-specific colour may also be shown with other species. Indeed, a species-specific enzyme reacting with the chromogenic substrate might be produced by distinct yeast species [27,28,29,30]. Furthermore, the variation in enzyme production among strains of the same Candida species could cause misidentification, as reported by other authors [26,31].
In our study, C. albicans (43.37%) was the most common yeast encountered in all clinical sources. This result is consistent with previous reports from symptomatic patients in Burkina Faso and other parts of the world [5,32,33,34,35,36]. In this study, Candida non-albicans species together represented a greater proportion than C. albicans did, meaning that in Burkina Faso, almost half of suspected candidiasis cases are caused by non-albicans species. Indeed, there is a rising prevalence of candidiasis caused by non-albicans Candida species worldwide [37,38,39]. The use of accurate diagnostic methods (MALDITOF MS and ITS sequencing) allowed the identification of uncommon species, including C. rugosa, C. mesorugosa, C. guilliermondii, and C. fermentati, not described before at the SSUH of Bobo-Dioulasso.
In this context, the accurate identification of Candida spp. and yeast-like organisms such as Trichosporon spp. is crucial for candidiasis management due to their variable innate susceptibility to antifungal drugs and the different clinical interpretative breakpoints of antifungals among Candida spp. Indeed, C. krusei is naturally resistant to fluconazole, C. glabrata is susceptibly dose-dependent to fluconazole, and Trichosporon spp. are resistant to echinocandins [7,40]. Azoles are the most currently used drugs used in Burkina Faso for the treatment of candidiasis, and yeast misidentification might jeopardise treatment outcomes because of inadequate antifungal drug administration or inappropriate posology.
Due to the high cost of more accurate microbiological tools, laboratories in resource-constrained settings might use accessible methods such as CCL together with other techniques to improve the accuracy of the mycological information delivered to the clinicians [41].
Finally, our results highlight the importance of implementing an internal quality system that ensures good manufacturing and training in the reading of chromogenic media in Burkina Faso. The use of another diagnostic method in addition to CCL in routine work is recommended, specifically in cases of invasive candidiasis. The deployment of a mycology reference centre in Burkina Faso, equipped with more sophisticated methods, could be an efficient alternative to help the management of complex cases in fungal pathology in national health centres.
Limitations
Our study had some limitations. Firstly, the small size of the study sample was a limitation, although chromogenic medium-detectable species account for the majority. Therefore, the results of this study could not be extrapolated to the entire population of Bobo-Dioulasso. Secondly, the CCL medium was prepared and read in routine practice conditions of the clinical laboratory, reflecting the real-life performance of this method. Finally, some parameters such as the reader’s experience could affect the performance of the test, as suggested by other authors [42].
5. Conclusions
In conclusion, this study assessed, for the first time, the performance of Chromogenic Candida Lab-Agar® for the identification of relevant clinical yeast isolates in the routine work of a clinical laboratory in Burkina Faso. The identification of pathogenic yeasts based on chromogenic media, in a resource-constrained setting, presents several limitations for identifying non-albicans Candida species (43.37% of misidentifications) that microbiologists and clinicians should consider. Around half of the yeast isolates of various clinical origins were Candida non-albicans, including uncommon species such as C. fermentati, C. mesorugosa, and Trichosporon spp. These facts underlie the potential discrepancies in antifungal treatment outcomes in a setting where azoles remained the main antifungal agent. Our results indicate the need to add a complementary test in the case of a non-albicans isolate identified via the CCL medium, specifically in the case of invasive candidiasis.
Author Contributions
S.N.D., S.B. and H.R.-V conceived and planned the experiments. S.N.D., B.B., and I.M. (Isidore Mandy) carried out the laboratory analysis. H.R.-V. and S.B. supervised the experiments. S.N.D. drafted the manuscript. I.W.Y., A.A., O.D. and I.M. (Isabel Montesinos) provided critical feedback and contributed to the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
We would like to thank the staff of the Laboratory department of Sourô Sanou University Hospital in Bobo Dioulasso, Burkina Faso, and the staff of the Microbiology department of Cliniques Universitaires Saint Luc, Brussels, Belgium.
Conflicts of Interest
The authors declare no conflicts of interests.
Abbreviations
| CCL | Chromogenic Candida Lab-Agar® |
| UCLouvain | Université Catholique de Louvain |
| ITS | Internal transcribed spacer |
| SDA | Sabouraud dextrose agar |
References
- Pappas, P.G.; Kauffman, C.A.; Andes, D.R.; Clancy, C.J.; Marr, K.A.; Ostrosky-Zeichner, L.; Reboli, A.C.; Schuster, M.G.; Vazquez, J.A.; Walsh, T.J.; et al. Clinical Practice Guideline for the Management of Candidiasis: 2016 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2016, 62, e1–e50. [Google Scholar] [CrossRef] [PubMed]
- Seyoum, E.; Bitew, A.; Mihret, A. Distribution of Candida Albicans and Non-Albicans Candida Species Isolated in Different Clinical Samples and Their in Vitro Antifungal Suscetibity Profile in Ethiopia. BMC Infect. Dis. 2020, 20, 231. [Google Scholar] [CrossRef] [PubMed]
- Sanata, B.; Salam, O.A.; Ibrahim, S.; Adama, Z.; Mamoudou, C.; Simplice, K.D.; Jacques, S.; Robert, G.T.; Christophe, H. Digestive Fungal Flora in Asymptomatic Subjects in Bobo-Dioulasso, Burkina Faso. Asian Pac. J. Trop. Biomed. 2014, 4, 659–662. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J.; Gibbs, D.L.; Newell, V.A.; Ellis, D.; Tullio, V.; Rodloff, A.; Fu, W.; Ling, T.A. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: A 10.5-Year Analysis of Susceptibilities of Candida Species to Fluconazole and Voriconazole as Determined by CLSI Standardized Disk Diffusion. J. Clin. Microbiol. 2010, 48, 1366–1377. [Google Scholar] [CrossRef] [PubMed]
- Sangaré, I.; Sirima, C.; Bamba, S.; Zida, A.; Cissé, M.; Bazié, W.W.; Sanou, S.; Dao, B.; Menan, H.; Guiguemdé, R.T. Prevalence of Vulvovaginal Candidiasis in Pregnancy at Three Health Centers in Burkina Faso. J. De Mycol. Médicale 2018, 28, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Alexander, B.D.; Johnson, M.D.; Pfeiffer, C.D.; Jiménez-Ortigosa, C.; Catania, J.; Booker, R.; Castanheira, M.; Messer, S.A.; Perlin, D.S.; Pfaller, M.A. Increasing Echinocandin Resistance in Candida Glabrata: Clinical Failure Correlates With Presence of FKS Mutations and Elevated Minimum Inhibitory Concentrations. Clin. Infect. Dis. 2013, 56, 1724–1732. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J.; Sheehan, D.J. Interpretive Breakpoints for Fluconazole and Candida Revisited: A Blueprint for the Future of Antifungal Susceptibility Testing. Clin. Microbiol. Rev. 2006, 19, 435–447. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Messer, S.A.; Hollis, R.J.; Boyken, L.; Tendolkar, S.; Kroeger, J.; Diekema, D.J. Variation in Susceptibility of Bloodstream Isolates of Candida Glabrata to Fluconazole According to Patient Age and Geographic Location in the United States in 2001 to 2007. J. Clin. Microbiol. 2009, 47, 3185–3190. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Messer, S.A.; Woosley, L.N.; Jones, R.N.; Castanheira, M. Echinocandin and Triazole Antifungal Susceptibility Profiles for Clinical Opportunistic Yeast and Mold Isolates Collected from 2010 to 2011: Application of New CLSI Clinical Breakpoints and Epidemiological Cutoff Values for Characterization of Geographic and Temporal Trends of Antifungal Resistance. J. Clin. Microbiol. 2013, 51, 2571–2581. [Google Scholar] [CrossRef]
- Borman, A.M.; Muller, J.; Walsh-Quantick, J.; Szekely, A.; Patterson, Z.; Palmer, M.D.; Fraser, M.; Johnson, E.M. Fluconazole Resistance in Isolates of Uncommon Pathogenic Yeast Species from the United Kingdom. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- Clark, A.E.; Kaleta, E.J.; Arora, A.; Wolk, D.M. Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry: A Fundamental Shift in the Routine Practice of Clinical Microbiology. Clin. Microbiol. Rev. 2013, 26, 547–603. [Google Scholar] [CrossRef] [PubMed]
- Freydiere, A.-M.; Guinet, R.; Boiron, P. Yeast Identification in the Clinical Microbiology Laboratory: Phenotypical Methods. Med. Mycol. 2001, 39, 9–33. [Google Scholar] [CrossRef] [PubMed]
- Kibwana, U.O.; Manyahi, J.; Kamori, D.; Mushi, M.; Mwandigha, A.M.; Majigo, M. Predominance of Non- Candida Albicans Species Oral Colonisation among Patients on Anticancer Therapy: Findings from a Cross-Sectional Study in Tanzania. BMJ Open 2023, 13, e070003. [Google Scholar] [CrossRef] [PubMed]
- Bernal, S.; Martín Mazuelos, E.; García, M.; Aller, A.I.; Martínez, M.A.; Gutiérrez, M.J. Evaluation of CHROMagar Candida Medium for the Isolation and Presumptive Identification of Species of Candida of Clinical Importance. Diagn. Microbiol. Infect. Dis. 1996, 24, 201–204. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; Volume 18, pp. 315–322. [Google Scholar] [CrossRef]
- Agarwal, S.; Manchanda, V.; Verma, N.; Bhalla, P. Yeast Identification in Routine Clinical Microbiology Laboratory and Its Clinical Relevance. Indian J. Med. Microbiol. 2011, 29, 172–177. [Google Scholar] [CrossRef]
- Bharathi, R. Comparison of Chromogenic Media with the Corn Meal Agar for Speciation of Candida. J. Pure Appl. Microbiol. 2018, 12, 1617–1622. [Google Scholar] [CrossRef]
- Robert, M.-G.; Cornet, M.; Hennebique, A.; Rasamoelina, T.; Caspar, Y.; Pondérand, L.; Bidart, M.; Durand, H.; Jacquet, M.; Garnaud, C.; et al. MALDI-TOF MS in a Medical Mycology Laboratory: On Stage and Backstage. Microorganisms 2021, 9, 1283. [Google Scholar] [CrossRef]
- Patel, R. A Moldy Application of MALDI: MALDI-ToF Mass Spectrometry for Fungal Identification. JoF 2019, 5, 4. [Google Scholar] [CrossRef]
- Ainscough, S.; Kibbler, C.C. An Evaluation of the Cost-Effectiveness of Using CHROMagar for Yeast Identification in a Routine Microbiology Laboratory. J. Med. Microbiol. 1998, 47, 623–628. [Google Scholar] [CrossRef]
- Souza, M.N.; Ortiz, S.O.; Mello, M.M.; Oliveira, F.d.M.; Severo, L.C.; Goebel, C.S. Comparison between Four Usual Methods of Identification of Candida Species. Rev. Inst. Med. Trop. Sao Paulo 2015, 57, 281–287. [Google Scholar] [CrossRef]
- Scharmann, U.; Kirchhoff, L.; Chapot, V.l.S.; Dziobaka, J.; Verhasselt, H.L.; Stauf, R.; Buer, J.; Steinmann, J.; Rath, P.-M. Comparison of Four Commercially Available Chromogenic Media to Identify Candida Albicans and Other Medically Relevant Candida Species. Mycoses 2020, 63, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Cetinkaya, Z.; Altindiş, M.; Aktepe, O.C.; Karabiçak, N. Comparison of different methods for the identification of Candida species isolated from clinical specimens. Mikrobiyol. Bul. 2003, 37, 269–276. [Google Scholar] [PubMed]
- Eraso, E.; Moragues, M.D.; Villar-Vidal, M.; Sahand, I.H.; González-Gómez, N.; Pontón, J.; Quindós, G. Evaluation of the New Chromogenic Medium Candida ID 2 for Isolation and Identification of Candida Albicans and Other Medically Important Candida Species. J. Clin. Microbiol. 2006, 44, 3340–3345. [Google Scholar] [CrossRef] [PubMed]
- Ilkit, M.; Hilmioglu, S.; Tasbakan, M.; Aydemir, S. Evaluation of Albicans ID2 and Biggy Agar for the Isolation and Direct Identification of Vaginal Yeast Isolates. J. Med. Microbiol. 2007, 56, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, K.; Ilkit, M.; Ates, A.; Turac-Bicer, A.; Demirhindi, H. Performance of Chromogenic Candida Agar and CHROMagar Candida in Recovery and Presumptive Identification of Monofungal and Polyfungal Vaginal Isolates. Med. Mycol. 2010, 48, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Sendid, B.; François, N.; Standaert, A.; Dehecq, E.; Zerimech, F.; Camus, D.; Poulain, D. Prospective Evaluation of the New Chromogenic Medium CandiSelect 4 for Differentiation and Presumptive Identification of the Major Pathogenic Candida Species. J. Med. Microbiol. 2007, 56 Pt 4, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Leena Sankari, S.; Mahalakshmi, K.; Naveen Kumar, V. Chromogenic Medium versus PCR–RFLP in the Speciation of Candida: A Comparative Study. BMC Res. Notes 2019, 12, 681. [Google Scholar] [CrossRef]
- Odds, F.C.; Bernaerts, R. CHROMagar Candida, a New Differential Isolation Medium for Presumptive Identification of Clinically Important Candida Species. J. Clin. Microbiol. 1994, 32, 1923–1929. [Google Scholar] [CrossRef]
- Ghelardi, E.; Pichierri, G.; Castagna, B.; Barnini, S.; Tavanti, A.; Campa, M. Efficacy of Chromogenic Candida Agar for Isolation and Presumptive Identification of Pathogenic Yeast Species. Clin. Microbiol. Infect. 2008, 14, 141–147. [Google Scholar] [CrossRef]
- Cooke, V.M.; Miles, R.J.; Price, R.G.; Midgley, G.; Khamri, W.; Richardson, A.C. New Chromogenic Agar Medium for the Identification of Candida Spp. Appl. Environ. Microbiol. 2002, 68, 3622–3627. [Google Scholar] [CrossRef]
- Zida, A.; Yacouba, A.; Bamba, S.; Sangare, I.; Sawadogo, M.; Guiguemde, T.; Kone, S.; Traore, L.K.; Ouedraogo-Traore, R.; Guiguemde, R.T. In Vitro Susceptibility of Candida Albicans Clinical Isolates to Eight Antifungal Agents in Ouagadougou (Burkina Faso). J. De Mycol. Médicale 2017, 27, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Ouanes, A.; Kouais, A.; Marouen, S.; Sahnoun, M.; Jemli, B.; Gargouri, S. Apport du milieu chromogène CHROMagar® Candida dans le diagnostic mycologique des levures. J. Mycol. Médicale 2013, 23, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Sule-Odu, A.O.; Akadri, A.A.; Oluwole, A.A.; Osinupebi, O.A.; Andu, B.A.; Akiseku, A.K.; Lawal, A.I. Vaginal Candida Infection in Pregnancy and Its Implications for Fetal Well-Being. Afr. J. Reprod. Health 2020, 24, 33–40. [Google Scholar] [PubMed]
- Risum, M.; Astvad, K.; Johansen, H.K.; Schønheyder, H.C.; Rosenvinge, F.; Knudsen, J.D.; Hare, R.K.; Datcu, R.; Røder, B.L.; Antsupova, V.S.; et al. Update 2016–2018 of the Nationwide Danish Fungaemia Surveillance Study: Epidemiologic Changes in a 15-Year Perspective. JoF 2021, 7, 491. [Google Scholar] [CrossRef] [PubMed]
- Gloria, A.; Marie, H.; Ashley, G. An Investigation of the Distribution of Candida Species in Genitourinary Candidiasis and Pelvic Inflammatory Disease from Three Locations in Ghana. Afr. J. Microbiol. Res. 2014, 8, 470–475. [Google Scholar] [CrossRef]
- Lass-Flörl, C.; Steixner, S. The Changing Epidemiology of Fungal Infections. Mol. Asp. Med. 2023, 94, 101215. [Google Scholar] [CrossRef] [PubMed]
- Sipsas, N.V.; Lewis, R.E.; Tarrand, J.; Hachem, R.; Rolston, K.V.; Raad, I.I.; Kontoyiannis, D.P. Candidemia in Patients with Hematologic Malignancies in the Era of New Antifungal Agents (2001–2007): Stable Incidence but Changing Epidemiology of a Still Frequently Lethal Infection. Cancer 2009, 115, 4745–4752. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J.; Turnidge, J.D.; Castanheira, M.; Jones, R.N. Twenty Years of the SENTRY Antifungal Surveillance Program: Results for Candida Species From 1997–2016. Open Forum Infect. Dis. 2019, 6 (Suppl. 1), S79–S94. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Boekhout, T.; Akova, M.; Meis, J.F.; Cornely, O.A.; Lortholary, O. ESCMID and ECMM Joint Clinical Guidelines for the Diagnosis and Management of Rare Invasive Yeast Infections. Clin. Microbiol. Infect. 2014, 20, 76–98. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Houston, A.; Coffmann, S. Application of CHROMagar Candida for Rapid Screening of Clinical Specimens for Candida Albicans, Candida Tropicalis, Candida Krusei, and Candida (Torulopsis) Glabrata. J. Clin. Microbiol. 1996, 34, 58–61. [Google Scholar] [CrossRef]
- Messeir, I.; M D S Abrantes, P.; W J Africa, C. Strengths and Limitations of Different Chromogenic Media for the Identification of Candida Species. Microbiology 2012, 2, 133–140. [Google Scholar] [CrossRef]
Figure 1.
Colour and appearance of quality control strains on chromogenic Candida Lab-Agar® medium after 48 h of incubation at 35 °C.
Figure 1.
Colour and appearance of quality control strains on chromogenic Candida Lab-Agar® medium after 48 h of incubation at 35 °C.
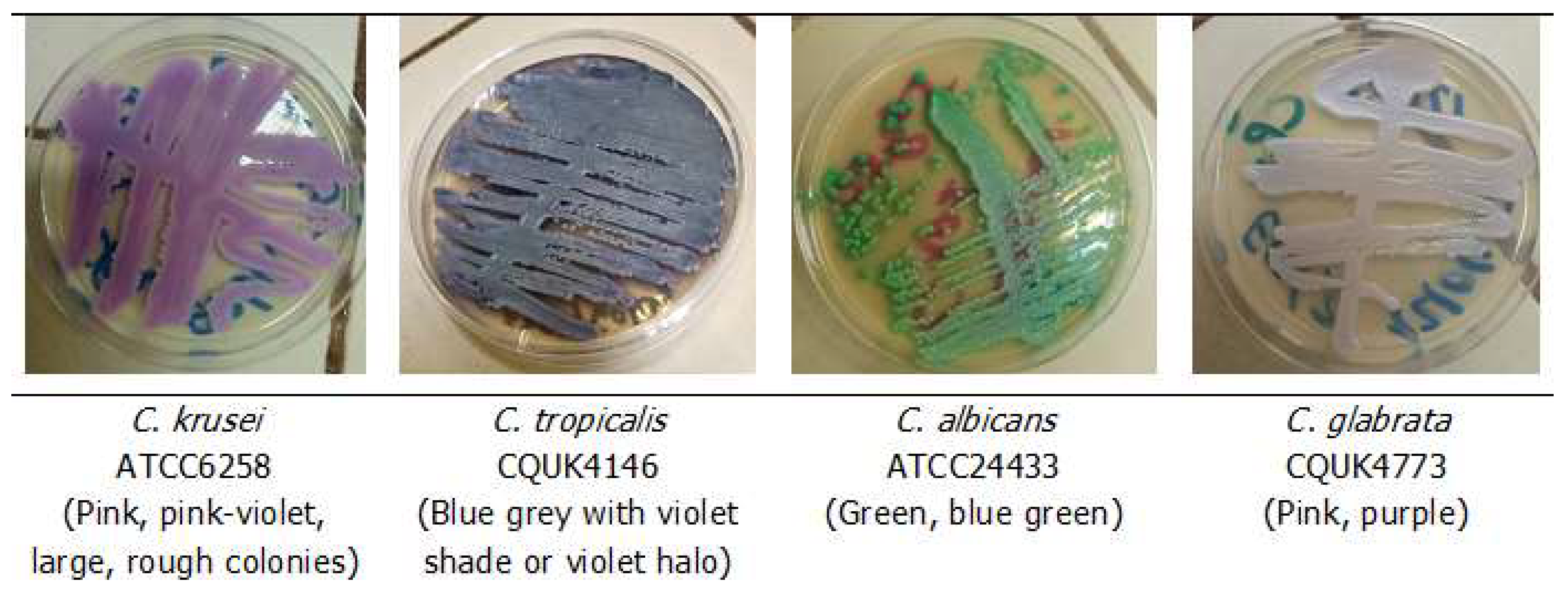
Table 1.
Distribution of 83 yeast isolates according to the species and clinical origins.
| Yeast Species * | Overall | Genital | Urine | Blood | ||||
|---|---|---|---|---|---|---|---|---|
| n = 83 | % | n = 48 | % | n = 33 | % | n = 2 | % | |
| C. albicans | 36 | 43.37 | 23 | 47.92 | 12 | 36.37 | 1 | 50.00 |
| C. krusei | 11 | 13.25 | 4 | 8.34 | 7 | 21.21 | - | - |
| C. glabrata | 10 | 12.05 | 8 | 16.67 | 2 | 6.06 | - | - |
| C. tropicalis | 6 | 7.22 | 2 | 4.17 | 4 | 12.12 | - | - |
| C. kefyr | 7 | 8.43 | 6 | 12.50 | 1 | 3.03 | - | - |
| C. lusitaniae | 2 | 2.41 | 1 | 2.08 | 1 | 3.03 | - | - |
| C. rugosa | 1 | 1.21 | - | - | 1 | 3.03 | - | - |
| C. mesorugosa | 2 | 2.41 | 1 | 2.08 | 1 | 3.03 | - | - |
| C. guilliermondii | 1 | 1.21 | - | - | 1 | 3.03 | - | - |
| C. fermentati | 3 | 3.61 | 1 | 2.08 | 1 | 3.03 | 1 | 50.00 |
| Cy. fabianii | 2 | 2.41 | 1 | 2.08 | 1 | 3.03 | - | - |
| T. asahii | 1 | 1.21 | - | - | 1 | 3.03 | - | - |
| T. inkin | 1 | 1.21 | 1 | 2.08 | - | - | - | - |
* Identification via MALDI-TOF MS and DNA sequencing. C.: Candida; Cy.: Cyberlindnera; and T.: Trichosporon.
Table 2.
Identification of 83 yeast isolates via CCL in comparison with the reference methods.
| Yeast Species * | CCL Identification | |||||
|---|---|---|---|---|---|---|
| C. albicans | C. glabrata | C. krusei | C. tropicalis | Undetermined | Total | |
| C. albicans | 34 | 0 | 0 | 0 | 2 | 36 |
| C. glabrata | 3 | 5 | 1 | 0 | 1 | 10 |
| C. krusei | 1 | 4 | 2 | 0 | 4 | 11 |
| C. tropicalis | 4 | 0 | 0 | 2 | 0 | 6 |
| C. kefyr | 2 | 2 | 3 | 0 | 0 | 7 |
| C. lusitaniae | 0 | 1 | 0 | 0 | 1 | 2 |
| C. rugosa | 0 | 0 | 0 | 0 | 1 | 1 |
| C. mesorugosa | 0 | 0 | 0 | 0 | 2 | 2 |
| C. guilliermondii | 0 | 1 | 0 | 0 | 0 | 1 |
| Cy. fabianii | 0 | 2 | 0 | 0 | 0 | 2 |
| C. fermentati | 1 | 2 | 0 | 0 | 0 | 3 |
| T. asahii | 1 | 0 | 0 | 0 | 0 | 1 |
| T. inkin | 1 | 0 | 0 | 0 | 0 | 1 |
| Total | 47 | 17 | 6 | 2 | 11 | 83 |
* Identification via MALDI-TOF MS and DNA sequencing. C.: Candida; Cy.: Cyberlindnera; and T.: Trichosporon.
Table 3.
Diagnostic performance of CCL in comparison to reference methods.
| Candida Species * | No. of CCL Concordant | No. of CCL Discordant | Sensitivity (%) | Specificity (%) | PPV ** (%) | NPV *** (%) |
|---|---|---|---|---|---|---|
| C. albicans (36) | 34 | 13 | 94.44 | 72.34 | 72.34 | 94.44 |
| C. glabrata (10) | 5 | 12 | 50.00 | 83.56 | 29.41 | 92.42 |
| C. krusei (11) | 2 | 4 | 18.18 | 94.44 | 33.33 | 88.31 |
| C. tropicalis (6) | 2 | 0 | 33.33 | 100 | 100 | 95.06 |
* Identification via MALDI-TOF MS and DNA sequencing. ** Positive Predictive value; *** negative Predictive value.
© 2024 Copyright by Authors. Licensed as an open access article using a CC BY 4.0 license.


