Afr. J. Parasitol. Mycol. Entomol. , 2(2), 16; doi:10.35995/ajpme02020016
Article
Low Circulation of Schistosoma haematobium x Schistosoma bovis Hybrid Species both in Perennial and Seasonal Schistosomiasis Transmission Areas in Senegal
1
EMR MINES Maladies Infectieuses, Négligées et Emergentes au Sud, Institut de Recherche pour le Développement, Campus International Institut de Recherche pour le Développement-Université Cheikh Anta Diop, Hann BP 1386, Dakar, Sénégal
2
Aix Marseille Université, Service de Santé des Armées, Assistance Publique-Hôpitaux de Marseille, RITMES, 19-21 Boulevard Jean Moulin, 13005 Marseille, France
3
Département de Biologie Animale, Faculté des Sciences et Techniques, Université Cheikh Anta Diop de Dakar, Dakar BP 5005, Sénégal
4
Département de Parasitologie-Mycologie, Unité de Formation et de Recherche des Sciences de la Santé, Université Gaston Berger, Saint-Louis BP 234, Sénégal
5
IHU Mediterranean Infection, 19-21 Bd Jean Moulin, 13005 Marseille, France
*
Corresponding author: thiamfatou545@gmail.com (F.T.); souleymane.doucoure@ird.fr (S.D.); Tel.: +221-77-516-54-85 (S.D.)
How to Cite: Thiam, F.; Sow, D.; Ranque, S.; Gaye, P.M.; L’Ollivier, C.; Parola, P.; Diamanka, A.; Doucoure, S.; Sokhna, C. Low circulation of Schistosoma haematobium x Schistosoma bovis hybrid species both in perennial and seasonal schistosomiasis transmission areas in Senegal. Afr. J. Parasitol. Mycol. Entomol., 2024, 2(2): 16; doi:10.35995/ajpme02020016.
Received: 19 August 2024 / Accepted: 28 November 2024 / Published: 23 December 2024
Abstract
:Background: In Northern Senegal, previous studies have shown a high occurrence of Schistosoma haematobium x S. bovis hybrids. This study aimed to update their circulation in perennial and seasonal transmission areas. Methods: Schistosoma eggs were collected in 2021 from 118 positive urine samples from 6- to 16-year-old children in the villages of Thiagar and Ndiathene, where schistosomiasis transmission is perennial, and in Niakhar and Khombole, with seasonal transmission. Dra1 Real-time PCR and RD PCR targeting mitochondrial (COX1) and nuclear DNA (ITS2) followed by sequencing were performed to characterize individual parasites. Results: Of the 1176 eggs obtained, only 998 with less than 33 ct at the Dra1 test were selected for RD PCR. Among these, 997 (99.9%) and 1 (0.1%) exhibited, respectively, S. haematobium and S. bovis patterns. Among the 998 COX1 amplicons, we obtained 997 sequences (99.9%) of S. haematobium and 1 (0.1%) of S. bovis (Ndiathene). Of the ITS2 sequences analyzed, 21 (2.1%) showed an S. haematobium x S. bovis hybrid sequence from Central Senegal. The complete genetic profiles showed that 976 (97.8%) and 22 (2.2%) sequences were S. haematobium and S. haematobium x S. bovis. Conclusion: Overall, hybrid prevalence was low and, for the first time, the circulation of S. haematobium x S. bovis hybrids was reported in human populations in Central Senegal. The prevalence of hybrids in the north is low compared with previous studies. The mapping of hybrid strains reinforces our knowledge of their circulation in Senegal.
Keywords:
schistosome hybrids; COX1/ITS2; perennial/seasonal transmission; Senegal1. Introduction
After malaria, schistosomiasis is the second most important parasitic disease in humans worldwide, affecting more than 250 million people with around 200 000 deaths every year [1]. Schistosomiasis is caused by flatworms of the genus Schistosoma. In sub-Saharan Africa, which accounts for around 90% of cases, most of the morbidity and mortality is caused by S. haematobium and S. mansoni, which are responsible for the urinary and intestinal forms of the disease. The S. haematobium clade includes three species that infect humans (S. haematobium, S. intercalatum and S. guineensis) and six species that can infect ruminants and/or rodents (S. bovis, S. curassoni, S. kisumuensis, S. leiperi, S. margrebowiei, and S. mattheei) [2].
However, all schistosome species share a two-host life cycle, with an asexual stage in a specific intermediate host, the snail, followed by a sexual stage in the definitive host, the mammal. During this second phase, inter-specific hybridization can occur between male and female worms of closely related schistosome species [3]. Hybrid strains S. haematobium X S. bovis and S. haematobium with the other sister species (S. curassoni and S. mattheei) have been identified in humans, following several studies in Africa using genetic methods including DNA sequencing, in Niger, Mali, Cote d’Ivoire, Malawi [4,5,6,7], and even in Corsica [8].
Hybrid strains can be identified using DNA markers based on the mitochondrial cytochrome oxidase subunit 1 gene (COX1) and the nuclear ribosomal internal transcribed spacer (ITS) region. The ITS1 and ITS2 markers can be inherited in a biparental manner while COX1 allows hybridization to be confirmed if the results obtained on species assignment are discordant. In addition, they can be identified with the presence of animal parasite alleles or haplotypes in humans, or when the ITS marker shows a heterozygous profile at specific mutation points [7,9,10]. In recent years, the emergence of hybrid strains has been noted in most endemic areas [6,7]. It is thought that hybrids have particular characteristics compared to pure S. haematobium strains. Indeed, they may have increased infectivity and fecundity, a faster maturation time, a much broader host spectrum, and therefore good adaptation [11,12]. It has been suggested that hybridization may reduce the efficacy of praziquantel, the only treatment available. However, to date, all the studies in endemic areas have shown the sensitivity of hybrid strains to praziquantel chemotherapy [3]. Thus, one of the major consequences of hybridization could be that the epidemiology of the disease is increasingly difficult to pin down because of the size of the animal reservoir. In endemic countries such as Senegal, S. haematobium x S. bovis hybrids have already been reported in both human and snail hosts, with prevalences ranging from 9 to 72% [3,9,13,14,15,16]. Most of these studies reported disparities in the distribution of hybrid strains, with frequencies varying from year to year and from place to place. Given this situation, which could complicate the epidemiology of the disease, it is necessary to have a fairly exhaustive map of the distribution of hybrids, especially in sites where transmission is intense with high prevalences. In Senegal, the majority of studies on hybrids have so far focused on the Senegal River area. The river watershed covers an area of 60,000 km, making it difficult to gain an exhaustive knowledge of the distribution and prevalence of hybrids. As a result, certain parts of the region remain unexplored to date. On the other hand, seasonal transmission areas have received very little attention when it comes to studying the presence of hybrids. The aim of this study was to contribute to a better understanding of the distribution of hybrids in the Senegal River delta and the seasonal transmission area in the center of the country.
2. Materials and Methods
2.1. Study Areas
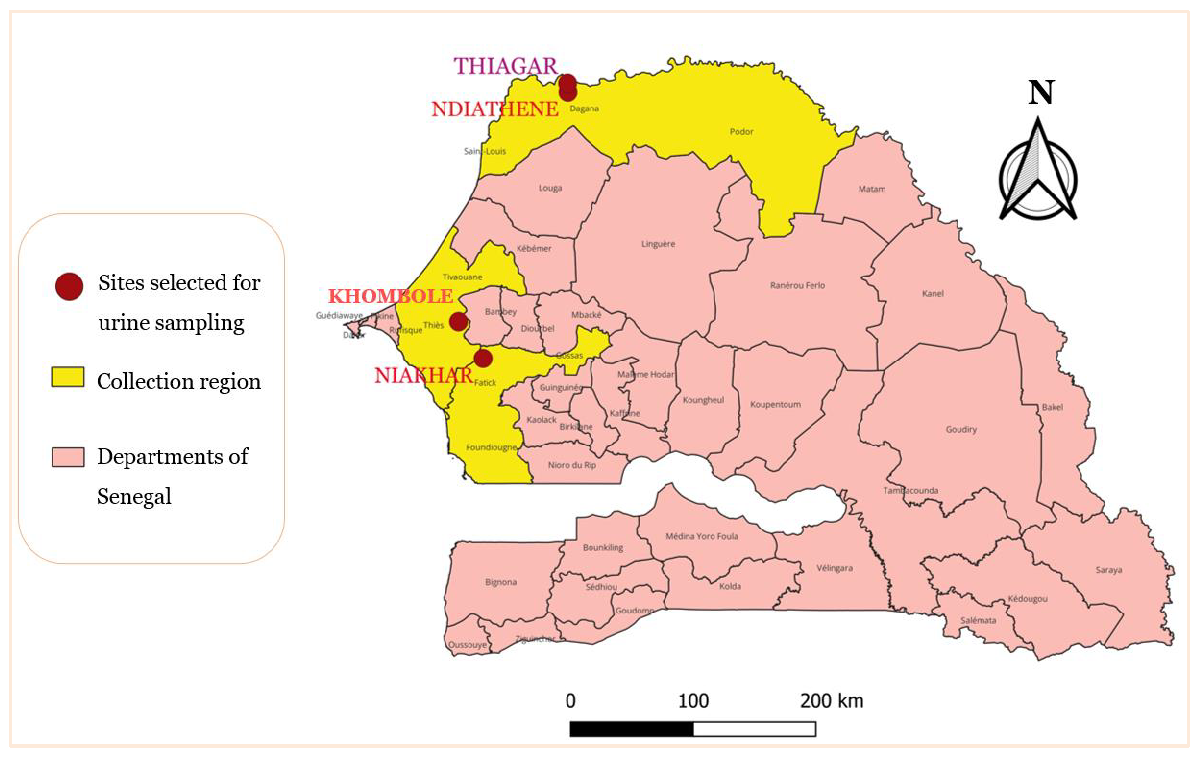
This study was conducted in central–western and northern Senegal where schistosomiasis trans-mission is temporary and permanent, respectively [17,18].In the northern part, in the Senegal River basin, schistosomiasis transmission occurs during the whole year and the prevalence of the disease can reach 90% in some localities [17]. In this area, the villages of Ndiathene (16°26 N and 15°52 W) and Thiagar (16°48 N and 15°85 W), at a distance of 1.8 km from one another, were selected (Figure 1). The two villages are located 20 km away from Richard-Toll and remain close to the Senegal River. In Thiagar, the population is estimated at about 1923 inhabitants, distributed in 216 concessions. Ndiathene has approximatively the same number of inhabitants (2000). These two zones are characterized by permanent pools frequented by the population for various activities (bathing, washing animals, etc.).
In the central–western part, the villages of Niakhar and Khombole were selected (Figure 1). Niakhar (14°30 N and 16°30 W) is located in the region of Fatick, 135 km east of Dakar. Khombole (14°46′ N and 16°41 W) is located in the region of Thies, 70 Km east of Dakar. Temporary ponds are observed only during the rainy season from July to November and are highly frequented by the population for domestic and recreative activities. Thus, the transmission of schistosomiasis occurs only during this period. In Niakhar, the transmission of the disease is well documented [18,19] while in Khombole, the transmission of schistosomiasis is poorly documented.
Figure 1.
Map of Senegal showing the location of the four urine collection sites: Ndiathene and Thiagar in the north, in the Saint-Louis region, and Niakhar and Khombole in the center, located, respectively, in the Fatick and Thies regions (produced with QGIS software).
Figure 1.
Map of Senegal showing the location of the four urine collection sites: Ndiathene and Thiagar in the north, in the Saint-Louis region, and Niakhar and Khombole in the center, located, respectively, in the Fatick and Thies regions (produced with QGIS software).
2.2. Study Populations
In this study, we used S. haematobium-positive urine samples collected from school-age children (6–16 years) in Thiagar, Ndiathene, Niakhar, and Khombole. The biological samples consisted of S. haematobium eggs obtained from these positive urine samples. In northern Senegal, in Thiagar and Ndiathene, the urine samples were collected during a clinical trial project on the efficacy of praziquantel in 2020 before the children took the drug (last treatment about 6 months ago). In Khombole, the urine samples were collected after an outbreak of urogenital schistosomiasis in 2021 before the children took the PZQ. In Niakhar, where the last mass treatment took place in 2016, the samples were collected during a schistosomiasis prevalence study in April 2018 [20]. For all the samples collected, informed consent was obtained from the legal tutor of minors.
2.3. Egg Collection
For each positive sample, an S. haematobium egg was isolated from urine, carefully homogenized, and centrifuged at 2000 rpm for 5 min before recovering and examining the pellets under a binocular loupe. Eggs were collected individually with a micropipette in a 3 µL volume and deposited on Whatman FTA Classic cards (GE Healthcare Life Sciences, Bucking-hamshire, UK) for molecular analysis. For each urine sample, between one (minimum) and twenty-four (maximum) schistosome eggs were recovered (Table 1), depending on the parasite load.
2.4. Molecular Analysis
2.4.1. Egg DNA Extraction
For DNA extraction, a 2 mm sample disk contain an individual egg was removed from the Whatman FTA® card using a metal unipolar punch forceps. The disk containing the egg was placed in an Eppendorf tube and incubated at 56 °C overnight with 180 μL of G2 lysis buffer (Qiagen Hilden, Germany) and 20 μL of proteinase K (Qiagen Hilden, Germany). The supernatant was collected in another tube for subsequent extraction with the EZ1 BioRobot extraction device (Qiagen Hilden, Germany) using the EZ1 DNA Tissue kit (Qiagen) according to the manufacturer’s recommendations. The eluate of each sample was recovered at the end (15 min) in a volume of 50 μL of Tris-EDTA buffer (Qiagen) and stored at −20 °C until use.
2.4.2. Real-Time PCR for Molecular Diagnosis of S. haematobium Group Species
For the Dra1 gene assay, all DNA extract samples from individual isolated eggs were run. A real-time PCR targeting the S. haematobium Dra1 sequence, 121 bp, constituting ~ 15% of the S. haematobium genome set [21], was carried with the following primers (forward 5′-GAT CTC ACC TAT CAG ACG AAAC-3′ and reverse 5′-TCA CAA CGA TAC GAC CAAC-3′) [22] and probe (5′-TGT TGG TGG AAG TGC CTG TTT CGCAA-3′) [23]. Dra1 PCR was performed within a 20 μL reaction mixture containing 5 μL of DNA, 3.5 μL of sterile ultrapure water, 0.5 μL of each primer, 0.5 μL of TaqManTM probe (Applied Bio-systems, Foster City, CA, USA), and 10 μL of Master Mix. The reaction was performed on a CFX96 thermal cycler (Bio-Rad, Marnes-la-Coquette, France), with the program consisting of an initial purification step of 2 min at 50 °C, followed by denaturation for 3 min at 95 °C, and hybridization for 40 cycles of 45 s at 60 °C; then, the sample was maintained at 4 °C.
DNA detection was expressed as the cycle threshold (Ct) value. The result was considered as positive if the cycle threshold (Ct) value was less than 35. At each assay, a negative out-of-model control (mix and sterile ultrapure water) and a positive S. haematobium group control (positive sample) were used.
2.4.3. Rapid Diagnostic PCR: Estimation of the Proportion of pure S. haematobium and Hybrid Population by Targeting COX1 Gene and ITS2 Region
All the positive Dra 1 qPCR samples were then used for the genetic characterization of the strains of S. haematobium, in search for the species infecting the cattle, in particular S. bovis and the hybrids, by a multilocus analysis of the mitochondrial gene COX and the nuclear region ITS [9,24].
Any sample with an S. bovis allele or haplotype or when the ITS2 marker showed a heterozygous profile (the presence of both S. haematobium and S. bovis alleles at a mutation site) was considered a hybrid between these two species [7,9,10].
-

- Identification of mitochondrial DNA (mtDNA COX1)
Each egg sample was molecularly characterized by multiplex PCR on COX1. We used species-specific primers to amplify the region in order to discriminate S. haematobium at 534 bp and S. bovis at 306 bp. The primers used were a single universal forward primer ShbmF, Asmit (5′-TTTTTTGGTCATCCTGAGGTGTAT-3′), with two reverse primers specific for the species S. haematobium, ShR (5′-TGATAATCAATGACCCTGCAATAA3′), and S. bovis, SbR (5′-CACAGGATCAGACAAACGAGTACC-3′) [24]. PCR amplification was performed in a 25 μL volume of reactions containing 12.5 μL of AmpliTaq Gold® 360 master mix (Applied Biosystems™, Waltham, MA, USA), 1.5 μL of the universal sense primer (ShbmF), 0.75 μL of each of the reverse primers (ShR and SbR), and 2 μL of DNA from each sample. PCR cycling conditions consisted of an initial denaturation step of 95 °C for 15 min, followed by 39 cycles of 95 °C for 30 s, 58 °C for 1 min, and 72 °C for 1 min, with a final extension of 7 min at 72 °C with a thermal cycler (Applied Biosystems, Foster City, CA, USA). The amplicons were then migrated for 25 min at 35 V on a 1.5% agarose gel stained with SYBR ® Safe (Invitrogen, Auckland, New Zealand) and visualized with a Gel Doc (Bio-Rad, Hercules, CA, USA).
-

- Amplification of the nuclear ITS2 region for species identification
Each positive sample with the COX1 gene was then amplified with the ITS2 region of the nuclear ribosomal DNA. DNA from each egg was performed in a 25 µL volume consisting of 12.5 µL Ampli Taq Gold buffer and 1 µL each of ITS2F (5′-GAA TTA ATG TGA ACT GCA TAC TGC TT-3′) and ITS2R (5′-TTC CTC CGC TTA TTG ATA TGC TT-3′) primers, as described by Quan and al. [25], 8.5 µL of ultrapure water, and 2 µL of genomic DNA from each sample, using the same thermal cycler. All PCR assays were performed using the following schedule, namely, an initial denaturation cycle at 95 °C for 15 min, followed by 39 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 1 min 30 s, and finally a cycle of 72 °C for 7 min, and then the products were also processed in the same manner as before. The results were used to assign each parasite a genetic signature based on haplotype–allele combinations.
2.4.4. Sequencing of Mitochondrial DNA (Cox1) and Nuclear rDNA (ITS2)
After amplification, products were purified using a Macherey Nagel plate (NucleoFast 96 PCR, Düren, Germany). All purified DNA products (partial COI mtDNA gene and ITS2 rDNA region) were then sequenced using the BigDye version 3.1 kit (Applied Biosystems, Foster City, CA, USA) using the original primers before being sequenced by the Sanger technique. BigDye consisted of amplifying the purified DNA from each egg in a final volume of 10 µL comprising 6 µL of mix and 4 µL of genomic DNA from each sample, using the same thermocycler. The mix consisted of one µL of BigDye, 1.5 µL of buffer, 0.5 µL of primer, and 3 µL of ultrapure water. All tests were performed using an initial denaturation cycle at 96 °C for 1 min, followed by 25 cycles of 96 °C for 10 s, 50 °C for 5 s, and 60 °C for 3 min before holding the sample at 15 °C. An ABI 3100 automated sequencer (Applied Biosystems) was used and the resulting sequences were manually edited and assembled using Chromas Pro software (version 5.1). The initial sequencing reads obtained from each isolate were aligned to the Egyptian strain of S. haematobium (JQ397368) available in the NCBI GenBank nucleotide data base, and only those that aligned correctly were considered for further analysis. The identity of the COX1 and ITS2 sequences from the individual eggs were confirmed by comparison to reference data and also using the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov). Nuclear sequence chromatograms (ITS2 rDNA) were used to search for polymorphic sites between S. haematobium and S. bovis. By reading the chromatograms, at each site where we had two overlapping peaks corresponding to the letters Y, R, or W, the IUPAC ambiguity symbols were used to determine the corresponding bases, and then we retained the letters when they indicated the genetic signatures of the two Schistosoma species, S. haematobium and S. bovis. The Cox1 and ITS2 genetic profiles were assigned to each individual egg to confirm species identity. Sequencing confirmed the pure or hybrid status of each egg sample.
With the sequences obtained, a multiple sequence alignment was constructed using clustalW1.6. The resulting fasta file was then converted to a mega-file format, and used to construct a phylogenetic tree based on Kimura’s 2-parameter method and 1000 bootstrap replicates as a measure of confidence interval in MEGA ver. 6. A phylogenetic tree for the gene Cox1 sequences was created using sequences available on the Genbank of S. haematobium from Senegal (FJ586241, JQ397330, JQ397331, JQ397333- JQ397336, JQ397338, JQ397340), Guinea Bissau (JQ397351), Mali (AY157209), Gambia (JQ397349), Cameroun (JQ397365), Nigeria (JQ397352), Egypt (JQ397368), S. bovis sequences from Senegal (AJ519521) and Tanzania (AY157212) and sequences of S. haematobium x S. bovis hybrids from 231 Corsica (KT354656, KT354658) and Benin (MW022141) (Figure S1).
2.5. Data Analysis
The data collected were entered in Excel. Variables were described in terms of numbers and percentages. We used the Chi-square to test whether the differences of the proportions of hybrids between villages were statistically significant. The probability values (p) < 0.05 were considered statistically significant.
2.6. Ethical Approval and Consent of Participants
The studies protocols were submitted to the Comité National d’Ethique pour la Recherche en Santé (CNERS) and received the ethical and scientific approval N° 000017/MSAS/DPRS/CNERS for Niakhar and N°19/MSAS/CNERS/Sec for Ndiathene and Thiagar. Urine samples were collected from children under the supervision of the Senegalese National Control Program of Schistosomiasis in Khombole district in April 2021. For children, consent was obtained from the parent individually.
3. Results
3.1. Sample Collection
In this study, we combined a total of 118 microscopy-positive S. haematobium urine samples. Of the 118 positive urine samples received, 39 samples were from the Niakhar area (Fatick), 62 samples comprised 42 from Ndiathene and 20 from Thiagar (Saint-Louis), and the remaining 17 were from Khombole. We individually isolated a total of 1176 eggs on Whatman FTA cards for molecular analysis, including 399 (33.9 %) eggs from the Ndiathene site, 161 (13.7%) from Thiagar, 388 (33%) from Niakhar, and 228 (19.4%) from the Khombole site (Table 1).
3.2. Real-Time PCR (Dra1 Gene)
A total of 1176 egg DNA extracts were tested for the DRA1 gene. PCR was positive in 1.130 samples, including 998 (84.9%) samples with a good signal (Ct values less than or equal to 33), and 132 (11.2%) samples with an average signal (Ct ranging from 33 to 35). On the other hand, PCR was negative in 46 (3.9%) samples (Ct values greater than 35). We selected the 998 positive samples (eggs) for subsequent analyses. These 998 eggs comprise 311 samples from Niakhar, 352 from Ndiathene, 137 from Thiagar, and 198 from Khombole (Table 1).
Table 1.
Number and distribution of urine samples included in the study, and number of S. haematobium eggs tested positive and negative by qPCR (Dra1).
Table 1.
Number and distribution of urine samples included in the study, and number of S. haematobium eggs tested positive and negative by qPCR (Dra1).
| Sites | No. of Positive Urine Samples (%) | No. of Eggs Isolated (%) | Mean Egg Value /Urine (Min-Max) | No. of Positive Eggs (Dra1) Ct < 33 33 < Ct < 35 | No. of Negative Eggs (Dra1) | |
|---|---|---|---|---|---|---|
| Ndiathene | 42 (35.6) | 399 (33.9) | 9 (2-15) | 352 | 36 | 11 |
| Thiagar | 20 (16.9) | 161 (13.7) | 8 (1-15) | 137 | 24 | 0 |
| Niakhar | 39 (33.1) | 388 (33) | 9 (1-20) | 311 | 45 | 32 |
| Khombole | 17 (14.4) | 228 (19.4) | 13 (1-24) | 198 | 27 | 3 |
| Total | 118 (100) | 1176 (100) | 998 (84.9) | 132 (11.2) | 46 (3.9) | |
3.3. Results of Amplification of the COX1 and ITS2 Regions
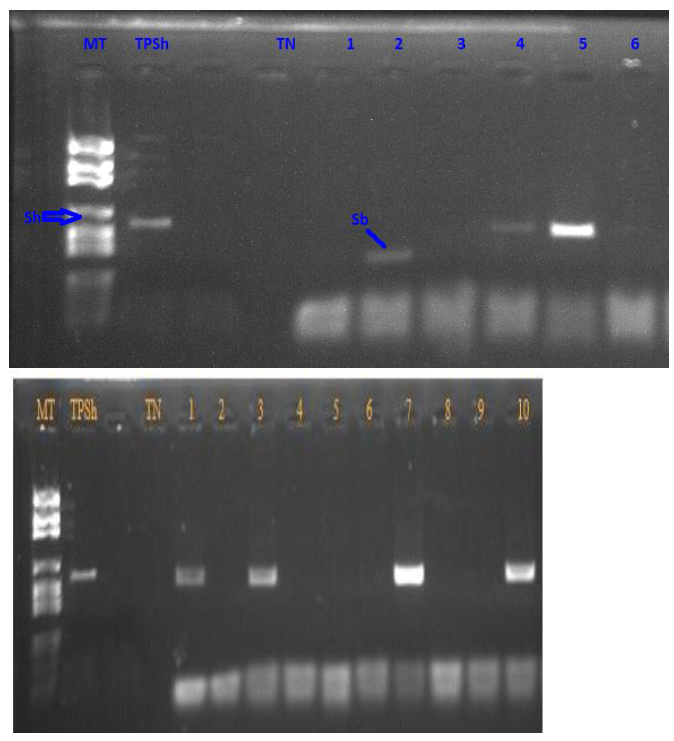
For each egg, after amplification, the migration of its DNA on an agarose gel showed either a 543 bp band, the size expected for S. haematobium, or a 306 bp band, corresponding to S. bovis (Figure 2). Only 1 egg sample with 306 bp was obtained while the remaining 997 egg samples were positive to 543 bp, giving thus a proportion of 0.1% (1/997) and 99.9% (997/998) for S. bovis and S. haematobium, respectively. The egg sample with an S. bovis profile was from the Ndiathene site where 352 individuals eggs were tested, giving therefore a proportion of 0.3% (1/352) for S. bovis and 99.7% (351/352) for S. haematobium. Then, for the other study sites (Thiagar, Niakhar, and Khombole), only S. haematobium was found based on the COX1 marker. All 998 samples amplified with the COX1 gene were then amplified with the ITS2 nuclear gene before being sequenced with the same genes to obtain the mitochondrial and nuclear genetic profile of each sample.
Figure 2.
Gel photos: Migration of eggs on agarose gel (543 bp band, the size expected for S. haematobium, and 306 bp band, corresponding to S. bovis).
Figure 2.
Gel photos: Migration of eggs on agarose gel (543 bp band, the size expected for S. haematobium, and 306 bp band, corresponding to S. bovis).
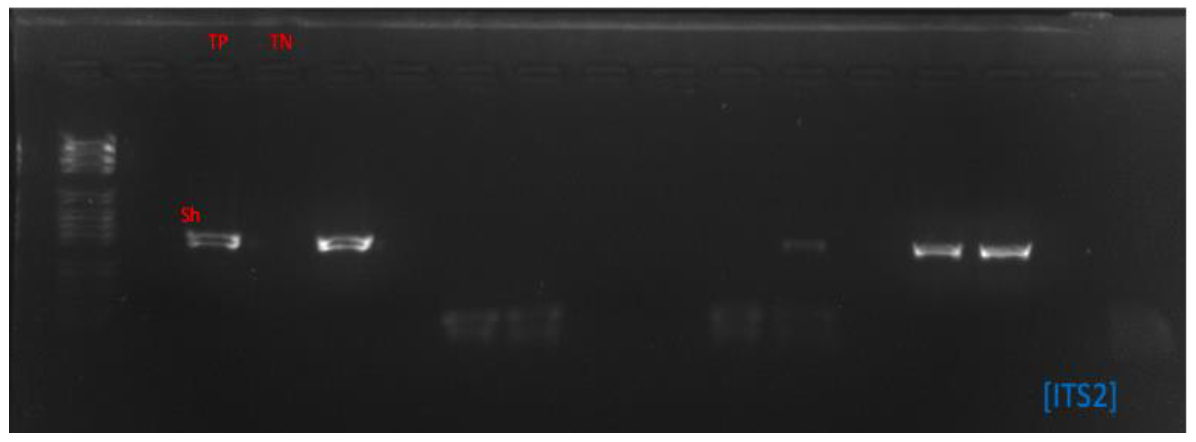
Bands (ITS2-PCR) of 468 pb were visualized on agarose gel. Sequencing with the same genes gives the mitochondrial and nuclear genetic profile of each sample (Figure 3).
Figure 3.
Gel photos of the migration of eggs on agarose gel with the ITS2 gene.
3.4. Results of Sequencing
3.4.1. COX1 Sequencing
The sequencing of the COX1 gene confirmed the results the PCR results. Thus, on the basis of COX1, only one egg sample (0.1%) with an S. bovis profile was obtained, and this was molecularly confirmed (GenBank: MT579445.1). This sample with an S. bovis profile was from the Ndiathene site (one in 352 eggs tested). In this area, we obtained a proportion of 0.3% (1/352) for S. bovis and 99.7% (351 out of 352) for S. haematobium. Then, for the other study sites (Thiagar, Niakhar, and Khombole), based only on the COX1 gene, we found only S. haematobium (GenBank: MK253577.1).
3.4.2. ITS2 Sequencing
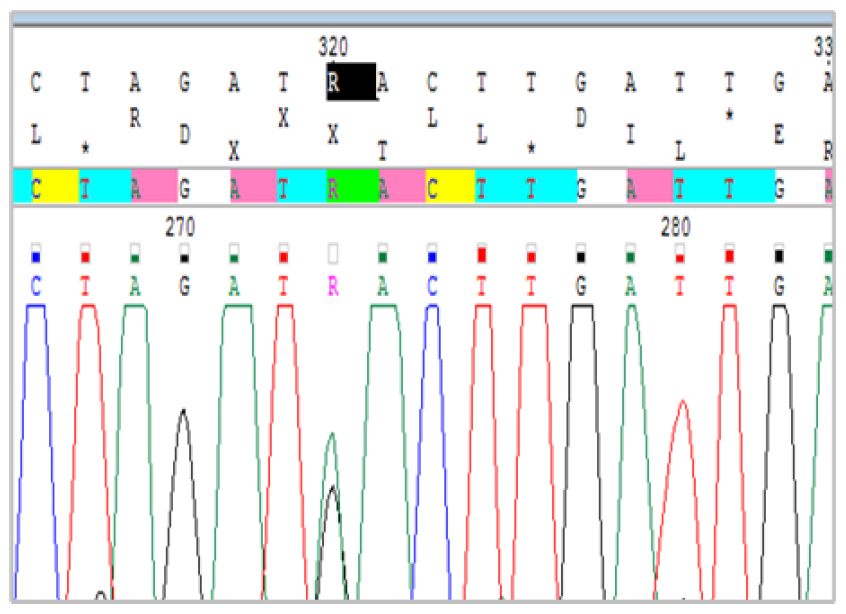
The sequencing of the ITS2 gene showed that 97.9% of the samples (977/998 eggs) were S. haematobium. The remaining 21 samples (2.1%), comprising 10 samples from Khombole and 11 from Niakhar (Table 2), showed a mixed sequence, characterized by the presence of two peaks on the Chromas Pro (Figure 4).
Figure 4.
Presence of two peaks (A and G, designated under the letter R) of different heights on the Chromas Pro.
Figure 4.
Presence of two peaks (A and G, designated under the letter R) of different heights on the Chromas Pro.
3.4.3. Results of Genotyping with COX1 and ITS2 Genes per Site
The nuclear (ITS2) and mitochondrial (COX1) identities of each individual egg were combined to produce a mitochondrial and nuclear genetic profile.
Of the 311 eggs from Niakhar, 300 (96.5%) had a pure S. haematobium genetic profile (COX1 and ITS2) and the remaining 11 (3.5%) had a COX1 S. haematobium profile and a mixed profile of S. haematobium and S. bovis with ITS2 (ShSb). Out of the 352 eggs from Ndiathene, 351 (99.7%) had a pure S. haematobium genetic profile and only 1 egg (0.3%) had a discordant S. bovis genetic profile with the mitochondrial gene (COX 1) and S. haematobium with the nuclear gene (ITS2). On the other hand, in the Thiagar area, all 137 eggs (100%) had a pure S. haematobium genetic profile. Of the 198 eggs from Khombole, 188 (94.9%) had a pure S. haematobium genetic profile and the remaining 10 (5.1%) a mixed profile of S. haematobium and S. bovis (COX1 S. haematobium and ITS2 S. haematobium x S. bovis) (Table 2).
In total, 22 (2.2%) hybrid eggs were sequenced in this study (11 from Niakhar, 10 from Khombole, and 1 from Ndiathene). They were collected from 13 urine samples (13 children), distributed as follows: 7 (7/39) from Niakhar, 5 (5/17) from Khombole, and 1 (1/42) from Ndiathene.
Table 2.
Prevalence of schistosome strains (S. haematobium and hybrid S. haematobium x S. bovis) for each site based on COX1 and ITS2 regions.
Table 2.
Prevalence of schistosome strains (S. haematobium and hybrid S. haematobium x S. bovis) for each site based on COX1 and ITS2 regions.
| Locality name | Number of | Pure genotypes (%) | Hybrid genotypes (%) | Number of hybrids (%) | |||
|---|---|---|---|---|---|---|---|
| Positive urine samples | Isolated eggs | Eggs with Ct < 33 | Sh x ShSh | Sh x ShSb | Sb x ShSh | ||
| Ndiathene | 42 | 399 | 352 | 351 (99.7) | 0 (0.0) | 1 (0.3) | 1 (0.3) |
| Thiagar | 20 | 161 | 137 | 137 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Niakhar | 39 | 388 | 311 | 300 (96.5) | 11 (3.5) | 0 (0.0) | 11 * (3.5) |
| Khombole | 17 | 228 | 198 | 188 (94.9) | 10 (5.1) | 0 (0.0) | 10 * (5.1) |
| TOTAL | 118 | 1176 | 998 | 976 (97.8) | 21 (2.1) | 1 (0.1) | 22 (2.2) |
Percentage (%) of genetic profiles obtained using the COX1 mitochondrial haploid gene (first two letters) and ITS2 diploid nuclear region (last four letters) per collection site. “Pure” Schistosoma haematobium (S. haematobium COX1 × S. haematobium ITS2: Sh × ShSh) and the two types of hybrids (type 2: S. bovis COX1 × S. haematobium ITS2 Sb × ShSh; and type1: S. haematobium COX1 × S. haematobium S. bovis ITS2 Sh × ShSb). * The 11 hybrid egg samples from Niakhar were obtained from 7 urine samples (children) and the 10 from Khombole from 5 urine samples (children).
The proportions of hybrids show statistically significant difference between Niakhar and Ndiathene (p = 0.0017) and between Ndiathene and khombole (p = 0.0001) but between Niakhar and khombole the difference is not significant (p = 0.422). This study shows a non-homogeneous distribution of hybrids between sites, with a statistically significant difference (p = 0.0016).
In summary, the 118 positive urine samples tested showed proportions of 11% (13/118) and 89% (105/118), respectively, of urine containing hybrid eggs and those containing only pure S. haematobium eggs. The children in whom hybrids were observed included 12 boys and a single girl living in the Niakhar area. We also found that all the children excreting hybrid eggs were in the 10–15 age group, except one, aged 7 (Table S1).
3.5. COX1 Phylogenies
The phylogenetic tree constructed (supplementary file Figure S1) shows the phylogenies of COX1 sequences (S. haematobium and S. bovis) obtained in this study. S. haematobium sequences from the four study sites were grouped with those from S. haematobium group 1. The S. bovis COX1 sequence was grouped with those of S. haematobium x S. bovis hybrids from Corsica.
4. Discussion
We determined the genetic profiles of individual eggs from S. haematobium-positive urine using PCR and sequencing techniques. The RD-PCR of maternally inherited haploid COX1 DNA was used to distinguish S. haematobium from S. bovis, and sequencing results for COX1 and ITS2 regions completed the genetic profiles. On the 1176 eggs, we identified 46 (3.9%) real-time PCR-negative egg samples and 132 (11.2%) samples with a Ct between 33 and 35 with the DRA1 gene. The agarose gel migration of these samples after amplification with the COX1 and ITS2 genes showed trace or no bands. This may be due to the small amount of DNA extracted. Indeed, comparative studies have shown that EZ1 can produce less amplifiable nucleic acid than other automated extractors [26]. Although these extractors can reduce handling time, their performance may depend on their ability to deliver amplifiable DNA [26], hence the importance of knowing the quantity and purity of DNA obtained by subsequent PCR [27,28] or target quantification [29,30,31,32]. We obtained 998 egg samples that showed the expected results in real-time PCR (Ct less than 33) and clearly visible bands on the agarose gel with the COX1 and ITS2 regions. These 998 egg samples were then successfully sequenced using the same genes. Thus, of the 998 samples sequenced, based on the two genes, we obtained a total of 22 (2.2%) hybrid eggs, including 11 from Niakhar, 10 from Khombole, and 1 from Ndiathene, from 13 different urine samples. This study therefore shows a predominance of pure strains of S. haematobium (97.8%). This study highlights for the first time the circulation of hybrid strains in human populations in the Niakhar and Khombole urogenital schistosomiasis seasonal transmission zone in Senegal. It also confirms earlier results showing the presence of hybrid parasites in the Bulinus snail population in Niakhar [19,33]. The circulation of hybrids is regularly documented in the north of the country, in the Senegal River basin. This study therefore improves the mapping of hybrid strains and adds to those that have already reported their presence in the temporary transmission zones of the Ferlo, in the east of the country in the Tambacounda region, and in the south in Kolda [3,16], suggesting a wide circulation of hybrid strains in Senegal. Comparing the four sampling sites, the percentage of hybrids varied from 0 to 5.05%, with the highest proportion found at Khombole. The distribution of hybrids therefore remains uneven between the populations sampled and between sites. This is consistent with previous studies carried out in Senegal (at Barkedji, Richard-Toll, Sinthiou Malem, etc.) showing highly variable hybrid rates between sites [3,13,14,16]. However, in this study, a low proportion of hybrid parasites was found. The percentages of hybrids found in Niakhar (2.25%) and Khombole (5.05%) are lower than those observed in Barkedji and Linguere, all characterized by the temporary transmission of urogenital bilharziasis, with hybrid percentages ranging from 10% in adults to 11% in children [16].
For the Niakhar site (Fatick), our study showed a percentage of S. haematobium x S. bovis hybrids of 2.25% out of 311 positive egg samples tested, showing that hybridization would easily occur in areas where the two species are co-endemic. Despite treatment, a variable level of transmission in different villages of over 34% has recently been reported [20]. Wider sampling is imperative to confirm these results, determine true prevalence, and show the possible role of these hybrids on disease transmission dynamics and treatment efficacy. At the Khombole site, a hybrid proportion of 5.05% was obtained out of 198 positive urine samples tested. Our study is the first to show both the presence of urogenital bilharziasis and hybrids between S. heamatobium and bovis in Khombole (Thies), located in the center of the country. Khombole is an area where livestock farming plays an important role; in addition, it is a quarrying area that contributes to the formation of temporary snail breeding sites, which are also frequented by children and animals. Further research is therefore needed to better understand the prevalence of the disease and the circulation of hybrid parasitic strains. Surprisingly, the prevalence of hybrid parasites was ultra-low at sites in the Senegal River basin, with only 0.3% at Ndiathene and no hybrid schistosome samples at Thiagar. These results drew our attention to the fact that hybrid prevalence can exceed 72% in this area [3,9,13,14,16]. This result could be explained first and foremost by a disparity in the distribution of hybrids. For example, the two villages are less than 2 km apart, but no hybrids were found in Thiagar. Although most of the work on hybrids in Senegal has been carried out in the northern part of the river basin, it should be noted that these earlier studies are not representative of the whole basin, and our study area, in the Senegal River delta, is still largely unexplored for bilharzia. On the other hand, the influence of socio-environmental conditions is an argument often put forward to explain the variation in hybrid frequency. It is therefore possible that the anthropogenic conditions conducive to hybridization are not identical throughout the Senegal River basin, which explains the low prevalence observed in this study. Leger and al. [16] have already highlighted the potential role of anthropogenic changes on hybridization rates in the Richard-Toll and Lac de Guiers areas, in contrast to Barkedji and Linguere (temporary pools). In Barkedji, the observed trend would also be linked to a lower prevalence of animal bilharziasis due to S. bovis than in the northern part of the country, as it is also clear that hybridization first implies that the geographical distributions of the two schistosome species and snail vectors overlap; otherwise, it cannot be a case of importation. It would be relevant to see whether the drop in prevalence in areas of perennial transmission [33] plays a role in the emergence of hybrids. Indeed, it is assumed that the prevalence of bilharzia may not be linked to hybrid strains [15], whereas in sites where S. bovis and S. haematobium are co-endemic, the relative importance of zoonotic transmission should increase as the disease in humans approaches elimination [34].
In the course of this study, type 1 (Niakhar and Khombole) and type 2 (Ndiathene) hybrids were found. These types of hybrid genetic profiles have already been encountered in northern Senegal and can be found in association within the same population [9,16]. Type 2 hybrid is the most frequently reported form. It has been found in Senegal from miracidia collected from infected patients [9], in Corsica [8], and also in Côte d’Ivoire on cercariae of intermediate snail hosts [35]. This result could testify to the ancient existence of hybrids at this site, as it has been shown that this form results from repeated backcrosses of S. haematobium x S. bovis hybrids with a pure strain of S. haematobium [2]. The hybrid of type 1 was more prominent in our study in contrast to previous studies in Senegal [3,9] and other countries such as Côte d’Ivoire [6]. These hybrids of type 1 generally correspond to those of the first generation [36]. This study focused only on school-age children and showed hybrid carriage in the 10–15 age group, as well as in younger children (7 years old). Adult sampling could provide additional information on the true proportion of hybrids in our study areas. Leger and al. [16] reported that adults can be well infected (up to 80%) with hybrids, hence the interest in incorporating all age groups in future studies for an accurate estimate of hybrid prevalence [37].
S. curassoni, also found in cattle and predominant in certain villages [16], can hybridize with S. bovis and/or S. haematobium [2]. Hybrids between S. bovis and S. curassoni have already been reported in Senegal [3,38]. Natural hybrids between S. haematobium and S. curassoni have also been notified in humans in Senegal and in Mali [3,5]. Hybrids between these three species have also been observed in Niger [2] and Mali [5]. However, we did not find these types of hybrids in this study, but we consider important to carry out a study of this kind in other transmission zones, using more genetic markers, in order to gain more in-depth knowledge of the types of hybrids and the different zones in which they circulate.
In Senegal, these ponds are still very popular with humans, but also with animals, especially cattle, and this facilitates encounters between these different species and the formation of hybrid schistosome progeny. The low prevalences found in this study could be explained by the hybrids’ sensitivity to PZQ. This would mean that the drug PZQ could still be considered effective in controlling this disease in areas where these hybrids circulate. In addition, it is important to note that although the samples were collected prior to treatment with PZQ, these villages are located in areas where annual mass treatments are carried out.
5. Conclusion
This study showed, for the first time, the presence of S. haematobium X S. bovis hybrids in the human population of Niakhar and Khombole located in the center of the country, whereas the prevalence reported in the north is low compared with previous studies. However, these results provide valuable information for the effective control and monitoring of urogenital bilharziasis. This study reveals a complex epidemiological situation and shows that the precise identification of schistosomes remains essential in order to know the genetic profile of the strains involved in transmission in each area. It would therefore be necessary to extend this study to gain a better mapping and knowledge of the impact of these hybridization events on the strategies implemented.
Supplementary Materials
The following supporting information can be downloaded at: https://ajpme.jams.pub/conversion/supplementary/01b2622dcc11fca0bfa3a41963bbba02. Figure S1: Phylogenetic tree with the gene COX1. This tree is derived from the maximum likelihood analysis of cox1 mitochondrial sequences from Schistosoma haematobium. The phylogeny of cox1 data was rooted with an S. mansoni haplotype (OU426849.1) (A) and S. curassoni (AJ519521.1) (B). All cox1 sequences were uploaded to the GenBank nucleotide data base. Clade support values for each node are maximum parsimony bootstrap percentages; Table S1: Urine samples with hybrid eggs. Number of eggs of pure S. haematobium and hybrids (S. haematobium x S. bovis) based on COX1 and ITS2 genes in each sample.
Author Contributions
T.F. conceived and designed the study; handled, analyzed, and interpreted the data; and drafted and revised the manuscript. S.D. conceived and designed the study, contributed materials, and revised the manuscript. R.S., L.C., and P.P. contributed reagents and materials, and revised the manuscript. G.P.M. handled, analyzed, and interpreted the data and revised the manuscript. D.A. revised the manuscript. D.S. conceived and designed the study, acquired funding, and drafted and revised the manuscript. S.C.: conceived and designed the study, contributed reagents and materials, acquired funding, and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research did not receive any specific funding grant. It was made possible through the support proved by the ARTS program (Research grants for a thesis in the South); the Institute of Research for Development (IRD), Dakar, Senegal; and the UGB/IRD Research Group on Neglected Tropical Diseases, Dakar, Senegal, and IHU Mediterranean Infection, Marseille, France.
Acknowledgments
We would like to thank the ARTS Program, IRD, and the IHU Mediterranean Infection for their support. We are grateful to all the technicians in the molecular biology and sequencing laboratory at the IHU (Marseille) for their help.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- WHO. Field Use of Molluscicides in Schistosomiasis Control Programmes: An Operational Manual for Programme Managers; World Health Organization: Geneva, Switzerland, 2017; Cataloguing at source; Available online: http://apps.who.int/iris (accessed on 20 June 2024).
- Leger, E.; Webster, J.P. Hybridizations within the genus Schistosoma: implications for evolution, epidemiology and control. Parasitology 2017, 144, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Webster, B.L.; Diaw, O.T.; Seye, M.M.; Webster, J.P.; Rollinson, D. Introgressive Hybridization of Schistosoma haematobium Group Species in Senegal: Species Barrier Break Down between Ruminant and Human Schistosomes. PloS Negl. Trop. Dis. 2013, 7, e2110. [Google Scholar] [CrossRef] [PubMed]
- Bremond, P.; Sellin, B.; Sellin, E.; Nameoua, B.; Labbo, R.; Théron, A.; Combes, C. Arguments for the modification of the genome (introgression) of the human parasite Schistosoma haematobium by genes from S. bovis, in Niger. C R. Acad. Sci. III 1993, 316, 667–70. [Google Scholar] [PubMed]
- Agniwo, P.; Boissier, J.; Sidibé, B.; Dembélé, L.; Diakité, A.; Niaré, D.S.; Akplogan, A.; Guindo, H.; Blin, M.; Dametto, S.; Ibikounlé, M. Genetic profiles of Schistosoma haematobium parasites from Malian transmission hotspot areas. Parasites Vectors 2023, 16, 263. [Google Scholar] [CrossRef] [PubMed]
- Angora, E.K.; Vangraefschepe, A.; Allienne, J.F.; Menan, H.; Coulibaly, J.T.; Meïté, A.; Raso, G.; Winkler, M.S.; Yavo, W.; Touré, A.O.; et al. Population genetic structure of Schistosoma haematobium and Schistosoma haematobium × Schistosoma bovis hybrids among school-aged children in Côte d’Ivoire. Parasite 2022, 29, 23, PMCID:PMC9074780. [Google Scholar] [CrossRef] [PubMed]
- Webster, B.L.; Alharbi, M.H.; Kayuni, S.; Makaula, P.; Halstead, F.; Christiansen, R.; Juziwelo, L.; Stanton, M.C.; LaCourse, E.J.; Rollinson, D.; et al. Schistosome interactions within the Schistosoma haematobium group, Malawi. Emerg. Infect. Dis. 2019, 25, 1245. [Google Scholar] [CrossRef]
- Boissier, J.; Moné, H.; Mitta, G.; Bargues, M.D.; Molyneux, D.; Mas-Coma, S. Schistosomiasis reaches Europe. Lancet Infect. Dis. 2015, 15, 757–758. [Google Scholar] [CrossRef]
- Huyse, T.; Webster, B.L.; Geldof, S.; Stothard, J.R.; Diaw, O.T.; Polman, K.; Rollinson, D. Bidirectional introgressive hybridization between cattle and human schistosome species. PLoS Pathog. 2009, 5, e1000571. [Google Scholar] [CrossRef]
- Pennance, T.; Archer, J.; Lugli, E.B.; Rostron, P.; Llanwarne, F.; Ali, S.M.; Amour, A.K.; Suleiman, K.R.; Li, S.; Rollinson, D.; et al. Development of a Molecular Snail Xenomonitoring Assay to Detect Schistosoma haematobium and Schistosoma bovis Infections in their Bulinus Snail Hosts. Molecules 2020, 25, 4011. [Google Scholar] [CrossRef]
- Southgate, V.R.; Tchuem Tchuenté, L.A.; Sène, M.; De Clercq, D.; Théron, A.; Jourdane, J.; Webster, B.L.; Rollinson, D.; Gryseels, B.; Vercruysse, J. Studies on the biology of schistosomiasis with a focus on the Senegal River basin. Memórias do Instituto Oswaldo Cruz. 2001, 96, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Webster, B.L.; Southgate, V.R. Compatibility of Schistosoma haematobium, S. intercalatum and their hybrids with Bulinus truncatus and B. forskalii. Parasitology 2003, 127, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Sene-Wade, M.; Marchand, B.; Rollinson, D.; Webster, B.L. Urogenital schistosomiasis and hybridization between Schistosoma haematobium and Schistosoma bovis in adults living in Richard-Toll, Senegal. Parasitology 2018, 145, 1723–1726. [Google Scholar] [CrossRef] [PubMed]
- Boon, N.A.; Mbow, M.; Paredis, L.; Moris, P.; Sy, I.; Maes, T.; Webster, B.L.; Sacko, M.; Volckaert, F.A.; Polman, K.; et al. No break in the barrier between human and bovine schistosome species in the Senegal River basin in the face of hybridization. Int. J. Parasitol. 2019, 49, 1039–1048. [Google Scholar] [CrossRef]
- Boon, N.A.M.; Van Den Broeck, F.; Faye, D.; Volckaert, F.A.M.; Mboup, S.; Polman, K.; Huyse, T. Barcoding hybrids: Heterogeneous distribution of Schistosoma haematobium × Schistosoma bovis hybrids across the Senegal River Basin. Parasitology 2018, 145, 634–645. [Google Scholar] [CrossRef]
- Léger, E.; Borlase, A.; Fall, C.B.; Diouf, N.D.; Diop, S.D.; Yasenev, L.; Catalano, S.; Thiam, C.T.; Ndiaye, A.; Emery, A.; et al. Prevalence and distribution of schistosomiasis in human, livestock, and snail populations in northern Senegal: A One Health epidemiological study of a multi-host system. Lancet Planet. Health 2020, 4, e330–e342. [Google Scholar] [CrossRef]
- Diop, B.; Sylla, K.; Kane, N.M.; Boh, O.K.; Guèye, B.; Ba, M.; Talla, I.; Mané, M.; Monteil, R.; Kinvi, B.; et al. Schistosomiasis control in Senegal: results of community data analysis to optimize praziquantel preventive chemotherapy intervention. Infect. Dis. Poverty 2023, 12, 106. [Google Scholar] [CrossRef]
- Senghor, B.; Diaw, O.T.; Doucoure, S.; Seye, M.; Diallo, A.; Talla, I.; Ba, C.T.; Sokhna, C. Impact of Annual Praziquantel Treatment on Urogenital Schistosomiasis in a Seasonal Transmission Focus in Central Senegal. PLoS Negl. Trop. Dis. 2016, 10, e0004557. [Google Scholar] [CrossRef] [PubMed]
- Gaye, P.M.; Doucoure, S.; Senghor, B.; Faye, B.; Goumballa, N.; Sembene, M.; L’Ollivier, C.; Parola, P.; Ranque, S.; Sow, D.; et al. Bulinus senegalensis and Bulinus umbilicatus Infestations of snails by the Schistosoma haematobium group in Niakhar, Senegal. Pathogens 2021, 10, 860, PMCID:PMC8308860. [Google Scholar] [CrossRef] [PubMed]
- Sow, D.; Sylla, K.; Dieng, N.M.; Senghor, B.; Gaye, P.M.; Fall, C.B.; Goumballa, N.; Diallo, A.; Ndiaye, J.L.A.; Parola, P.; et al. Molecular diagnosis of urogenital schistosomiasis in pre-school children, school-aged children and women of reproductive age at community level in central Senegal. Parasit. Vectors 2023, 6, 43, PMCID:PMC9887789. [Google Scholar] [CrossRef] [PubMed]
- Hamburger, J.; He-Na.; Abbasi, I.; Ramzy, R.M.; Jourdane, J.; Ruppel, A. Polymerase chain reaction assay based on a highly repeated sequence of Schistosoma haematobium: a potential tool for monitoring schistosome-infested water. Am. J. Trop. Med. Hyg. 2001, 65, 90711. [Google Scholar] [CrossRef] [PubMed]
- Ibironke, O.A.; Phillips, A.E.; Garba, A.; Lamine, S.M.; Shiff, C. Diagnosis of Schistosoma haematobium by detection of specific DNA fragments from filtered urine samples. Am. J. Trop. Med. Hyg. 2011, 84, 998–1001. [Google Scholar] [CrossRef]
- Cnops, L.; Soentjens, P.; Clerinx, J.; Van Esbroeck, M.A. Schistosoma haematobium-specific real-time PCR for diagnosis of urogenital schistosomiasis in serum samples of international travelers and migrants. PLoS Negl. Trop. Dis. 2013, 7, e2413, PMCID:PMC3757062. [Google Scholar] [CrossRef] [PubMed]
- Webster, B.L.; Rollinson, D.; Stothard, J.R.; Huyse, T. Rapid diagnostic multiplex PCR (RD-PCR) to discriminate between Schistosoma haematobium and S. bovis. J. Helminthol. 2010, 84, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.H.; Choi, I.W.; Ismail, H.A.; Mohamed, A.S.; Jeong, H.G.; Lee, J.S.; Hong, S.T.; Yong, T.S.; Cha, G.H.; Lee, Y.H. Genetic Diversity of Schistosoma haematobium Eggs Isolated from Human Urine in Sudan. Korean J. Parasitol. 2015, 53, 271–7, PMCID:PMC4510678. [Google Scholar] [CrossRef] [PubMed]
- Dundas, N.; Leos, N.K.; Mitui, M.; Revell, P.; Rogers, B.B. Comparison of automated nucleic acid extraction methods with manual extraction. J. Mol. Diagn. 2008, 10, 311–6, PMCID:PMC2438199. [Google Scholar] [CrossRef] [PubMed]
- Loens, K.; Bergs, K.; Ursi, D.; Goossens, H.; Ieven, M. Evaluation of NucliSens easyMAG for automated nucleic acid extraction from various clinical samples. J. Clin. Microbiol. 2007, 45, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Petrich, A.; Mahony, J.; Chong, S.; Broukhanski, G.; Gharabaghi, F.; Johnson, G.; Louie, L.; Luinstra, K.; Willey, B.; Akhaven, P.; et al. Multicenter comparison of nucleic acid extraction methods for the detection of severe acute respiratory syndrome coronavirus RNA in stool samples. J. Clin. Microbiol. 2006, 44, 2681–2688. [Google Scholar] [CrossRef]
- Beuselinck, K.; van Ranst, M.; van Eldere, J. Automated extraction of viral pathogen RNA and DNA for high-throughput real-time quantitative PCR. J. Clin. Microbiol. 2005, 43, 5541–5546. [Google Scholar] [CrossRef]
- Schuurman, T.; van Breda, A.; de Boer, R.; Kooistra-Smid, M.; Beld, M.; Savelkoul, P.; Boom, R. Reduced PCR sensitivity due to impaired DNA recovery with MagNA Pure LC total nucleic acid isolation kit. J. Clin. Microbiol. 2005, 43, 4616–4622. [Google Scholar] [CrossRef]
- Tang, YW.; Sefers, S.E.; Li, H.; Kohn, D.J.; Procop, G.W. Comparative evaluation of three commercial systems for nucleic acid extraction from urine samples. J. Clin. Microbiol. 2005, 43, 4830–4833. [Google Scholar] [CrossRef]
- Wilson, D.; Yen-Lieberman, B.; Reischl, U.; Warshawsky, I.; Procop, G.W. Comparison of five methods for extracting Legionella pneumophila from respiratory samples. J. Clin. Microbiol. 2004, 42, 5913–5916. [Google Scholar] [CrossRef]
- Senghor, B.; Webster, B.; Pennance, T.; Sène, M.; Doucoure, S.; Sow, D.; Sokhna, C. Molecular characterization of schistosome cercariae and their Bulinus snail hosts from Niakhar, a seasonal transmission focus in central Senegal. Curr. Res. Parasitol. Vector Borne Dis. 2023, 3, 100114, PMCID:PMC9941053. [Google Scholar] [CrossRef] [PubMed]
- Borlase, A.; Rudge, J.W.; Leger, E.; Diouf, N.D.; Fall, C.B.; Diop, S.D.; Catalano, S.; Sène, M.; Webster, J.P. Spillover, hybridization, and persistence in schistosome transmission dynamics at the human-animal interface. Proc. Natl. Acad. Sci. USA 2021, 118, e2110711118, PMCID:PMC8521685. [Google Scholar] [CrossRef] [PubMed]
- Tian-Bi, Y.T.; Webster, B.; Konan, C.K.; Allan, F.; Diakité, N.R.; Ouattara, M.; Salia, D.; Koné, A.; Kakou, A.K.; Rabone, M.; et al. Molecular characterization and distribution of Schistosoma cercariae collected from naturally infected bulinid snails in northern and central Côte d’Ivoire. Parasit. Vectors 2019, 12, 117, PMCID:PMC6423847. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.K. Modification des traits d’histoire de vie au cours de l’hybridation et analyse des mécanismes moléculaires sous-jacents chez les parasites plathelminthes du genre Schistosoma. Doctoral Dissertation, Université de Perpignan, Perpignan, France, 2018. [Google Scholar]
- Toor, J.; Turner, H.C.; Truscott, J.E.; Werkman, M.; Phillips, A.E.; Alsallaq, R.; Medley, G.F.; King, C.H.; Anderson, R.M. The design of schistosomiasis monitoring and evaluation programmes: The importance of collecting adult data to inform treatment strategies for Schistosoma mansoni. PLoS Negl. Trop. Dis. 2018, 12, e0006717, PMCID:PMC6175503. [Google Scholar] [CrossRef] [PubMed]
- Leger, E.; Garba, A.; Hamidou, A.A. Introgressed animal schistosomes Schistosoma curassoni and S bovis naturally infecting humans. Emerg Infect Dis. 2016, 22, 2212–2214. [Google Scholar] [CrossRef]
© 2024 Copyright by Authors. Licensed as an open access article using a CC BY 4.0 license.

